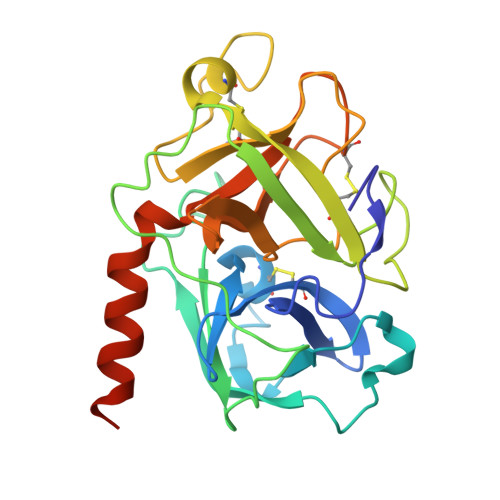
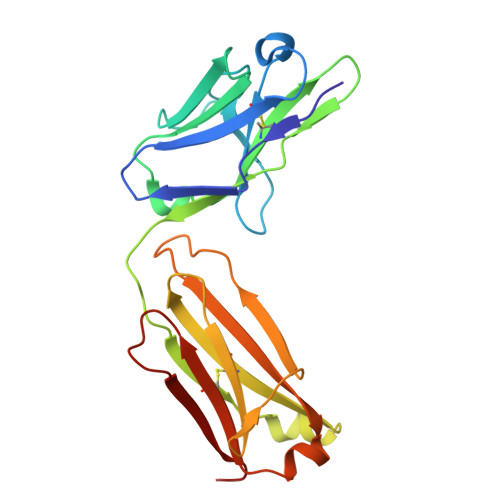
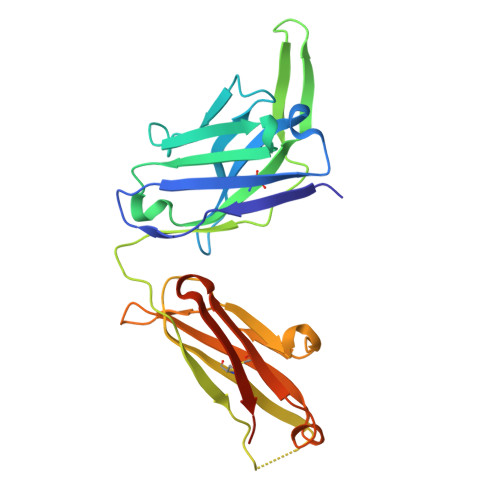
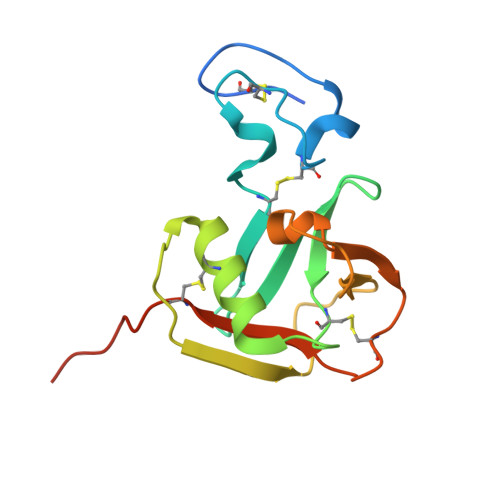
The crystal structure of coronavirus RBD-TMPRSS2 complex provides basis for the discovery of therapeutic antibodies.
Zhao, Z., Yang, Q., Liu, X., Li, M., Duan, Y., Du, M., Zhou, A., Liu, H., He, Y., Wang, W., Lu, Y., Zhang, X., Wang, H., Yang, X., Zhang, H., Chen, X., Rao, Z., Yang, H.(2025) Nat Commun 16: 6636-6636
- PubMed: 40681508
- DOI: https://doi.org/10.1038/s41467-025-62023-2
- Primary Citation of Related Structures:
9JCX, 9JCY, 9JD0, 9JD1, 9U8G - PubMed Abstract:
HCoV-HKU1, one of seven human coronaviruses (HCoVs) that have harmful effects on human health, accounts for a substantial portion of common cold cases and can cause severe respiratory diseases in certain populations. Currently, effective antiviral treatments against this virus are limited. Recently, TMPRSS2, a host protease long acknowledged for its role in priming the spike proteins of various CoVs and promoting viral entry, was identified as a functional receptor for HCoV-HKU1, opening an avenue for anti-HCoV-HKU1 therapy development. In this study, we elucidate the detailed molecular mechanism underlying the interaction between the HCoV-HKU1 receptor-binding domain (RBD) and TMPRSS2 via crystallography. Guided by these structural insights, we successfully develop two types of therapeutic antibodies against HCoV-HKU1. The first type neutralizes the RBD, potently disrupting its interaction with TMPRSS2 and preventing viral infection. The second type targets TMPRSS2, inhibiting its enzymatic activity and/or interfering with its binding to the RBD. The latter demonstrates broad-spectrum anti-CoV activity, as the enzymatic activity of TMPRSS2 is crucial for both HCoV-HKU1 infection and other CoV infections. Our findings provide crucial structural insights into the recognition of TMPRSS2 by HCoV-HKU1 and offer promising antibody-based strategies for combating HCoV-HKU1 and other CoV infections.
Organizational Affiliation:
Shanghai Institute for Advanced Immunochemical Studies and School of Life Science and Technology, ShanghaiTech University, Shanghai, China.