Alcaligenes faecalis Aio B C65F-C80G bound to Sb oxyanion
Engrola, F., Romao, M.J., Correia, M., Santos-Silva, T.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Arsenite oxidase subunit AioA | 822 | Alcaligenes faecalis | Mutation(s): 0 Gene Names: aioA, aoxB, asoA EC: 1.20.9.1 | 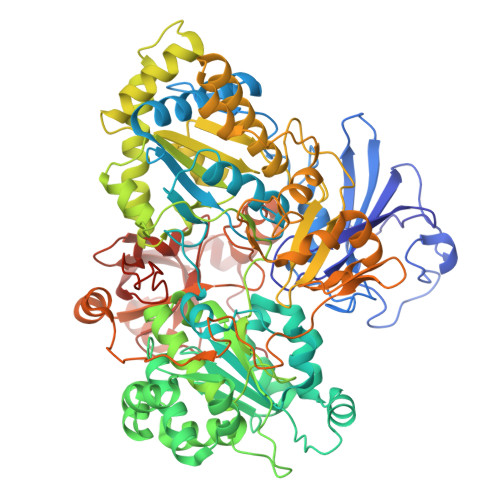 | |
UniProt | |||||
Find proteins for Q7SIF4 (Alcaligenes faecalis) Explore Q7SIF4 Go to UniProtKB: Q7SIF4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7SIF4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Arsenite oxidase subunit AioB | 134 | Alcaligenes faecalis | Mutation(s): 0 Gene Names: aioB, aoxA, asoB EC: 1.20.9.1 | 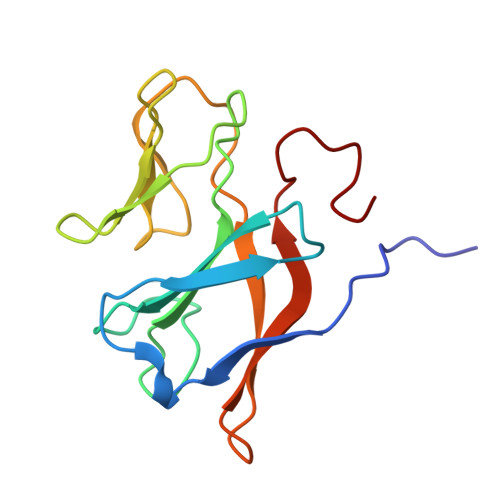 | |
UniProt | |||||
Find proteins for Q7SIF3 (Alcaligenes faecalis) Explore Q7SIF3 Go to UniProtKB: Q7SIF3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7SIF3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Arsenite oxidase subunit AioB | 135 | Alcaligenes faecalis | Mutation(s): 0 Gene Names: aioB, aoxA, asoB EC: 1.20.9.1 | 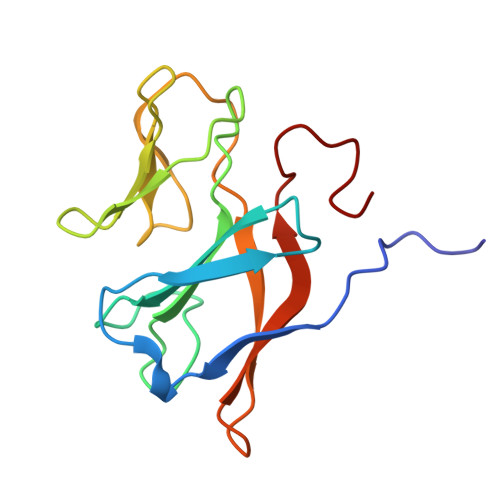 | |
UniProt | |||||
Find proteins for Q7SIF3 (Alcaligenes faecalis) Explore Q7SIF3 Go to UniProtKB: Q7SIF3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7SIF3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Arsenite oxidase subunit AioA | 824 | Alcaligenes faecalis | Mutation(s): 0 Gene Names: aioA, aoxB, asoA EC: 1.20.9.1 | 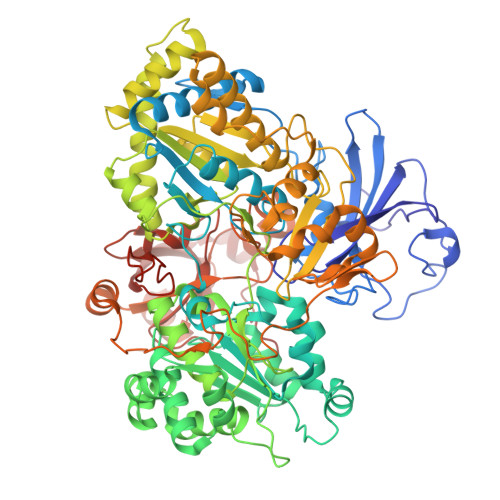 | |
UniProt | |||||
Find proteins for Q7SIF4 (Alcaligenes faecalis) Explore Q7SIF4 Go to UniProtKB: Q7SIF4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7SIF4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Arsenite oxidase subunit AioA | 823 | Alcaligenes faecalis | Mutation(s): 0 Gene Names: aioA, aoxB, asoA EC: 1.20.9.1 |  | |
UniProt | |||||
Find proteins for Q7SIF4 (Alcaligenes faecalis) Explore Q7SIF4 Go to UniProtKB: Q7SIF4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7SIF4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 13 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MGD (Subject of Investigation/LOI) Query on MGD | AA [auth C] BA [auth C] FB [auth G] GB [auth G] I [auth A] | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE C20 H26 N10 O13 P2 S2 VQAGYJCYOLHZDH-ILXWUORBSA-N |  | ||
| F3S (Subject of Investigation/LOI) Query on F3S | DA [auth C], HB [auth G], L [auth A], QA [auth E] | FE3-S4 CLUSTER Fe3 S4 FCXHZBQOKRZXKS-UHFFFAOYSA-N |  | ||
| 1PE Query on 1PE | HA [auth C], IA [auth C], PB [auth G], Q [auth A] | PENTAETHYLENE GLYCOL C10 H22 O6 JLFNLZLINWHATN-UHFFFAOYSA-N |  | ||
| FES (Subject of Investigation/LOI) Query on FES | DB [auth F], NA [auth D], VB [auth H], Z [auth B] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| P4G Query on P4G | EA [auth C] | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE C8 H18 O3 RRQYJINTUHWNHW-UHFFFAOYSA-N |  | ||
| PGE Query on PGE | T [auth A] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| PG0 Query on PG0 | OA [auth D] | 2-(2-METHOXYETHOXY)ETHANOL C5 H12 O3 SBASXUCJHJRPEV-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | AB [auth E] FA [auth C] GA [auth C] JB [auth G] KB [auth G] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| 4MO (Subject of Investigation/LOI) Query on 4MO | CA [auth C], EB [auth G], K [auth A], TA [auth E] | MOLYBDENUM(IV) ION Mo ZIKKVZAYJJZBGE-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | U [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | IB [auth G] JA [auth C] MA [auth D] MB [auth G] NB [auth G] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| NA Query on NA | CB [auth E], LA [auth C], TB [auth G], X [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| O (Subject of Investigation/LOI) Query on O | BB [auth E], KA [auth C], SB [auth G], W [auth A] | OXYGEN ATOM O XLYOFNOQVPJJNP-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 90.164 | α = 97.58 |
| b = 108.797 | β = 90.04 |
| c = 117.109 | γ = 96.18 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XDS | data scaling |
| REFMAC | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Fundacao para a Ciencia e a Tecnologia | Portugal | -- |